Quantifying glycosylation of HBsAg (Glyco-S/S):
To measure amounts of glycosylation of HBsAg

An analysis service using a new proprietary ELISA system for the first time measuring the extent of N-glycosylated S-HBs [Glyco-S ELISA].
Applicable Genotype: A, B, C and D
Overview
HBV virion is covered by envelope protein (HBsAg). HBsAg is modified with N- and O-glycan in subtype-specific manner (1). S-HBsAg is the most abundant and forms non-infectious particles. About half of S-HBsAgs is modulated by N-glycan and the rest is non-glycosylated. Infectious HBV particles contain S-, M-, and L-HBsAg and contains HBV DNA as well as core proteins. PreS2 domain in M-HBsAg is highly O-glycosylated, but not in L-HBsAg (2). Glycosylation of HBsAg is required for HBV formation and secretion in HBV life cycle (3). Furthermore, glycosylation of HBV can lead immune evasion by inhibiting recognition of antibody, antigen presenting molecules, and innate immune receptors (4). Thus, measuring glycosylation of HBsAg can reflect different pathological conditions of HBV patients, which is not revealed by qHBsAg measurement. For this purpose, we established a proprietary ELISA system, in which glycosylation of intact HBV can be quantified.
Glyco-S ELISA system can measure HBsAg glycosylation of genotype A, B, C, and D as far as we tested.
Quantifying glycosylation of HBsAg (Glyco-S/S) of HBV particles:
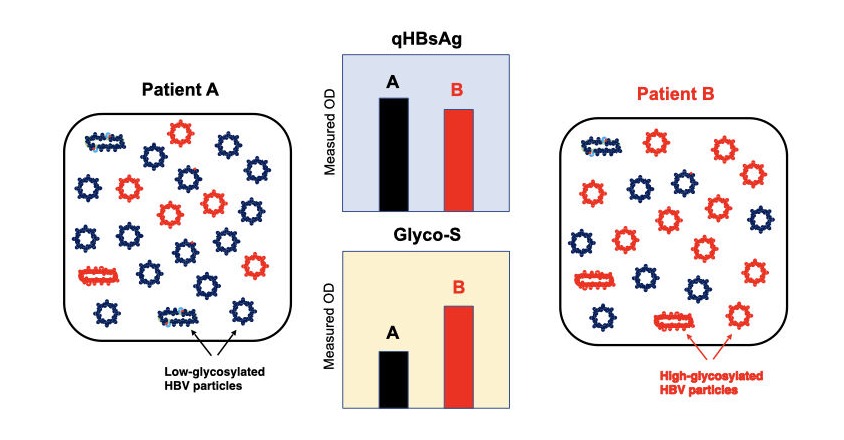
Composition of HBV particles in each patient is different like patient A and B. Amount of qHBsAg of patient A is comparable with that of B. But, Glyco-S of patient A is lower than that of B who has highly glycosylated HBV particles (red).
Our contract service covers clinical and non-clinical specimens:
- Sera from HBV patients of various status
- Sera containing HBV particles from experimental animals
- Cell culture media from primary human hepatocytes infected with HBV.
- HBV particles containing different glycosylation caused by mutations
For details and ordering this contract service, please feel free to contact us.
References:
1. Schmitt et al. (2004) J Gen Virol 85:2045-2053.
2. Angata et al. (2021) Biochim Biophys Acta Gen Subj. 1866:130020
3. Ouchida T et al. (2021) Viruses 13: 1860.
4. Dobrica et al. (2020) Cells 9:1404
Quantifying HBsAgGi:
To measure levels of HBsAg glycan isomer
An analysis service using HBsAgGi ELISA system for the first time measuring O-glycosylated M-HBsAg.
Applicable Genotype: B and C

Overview
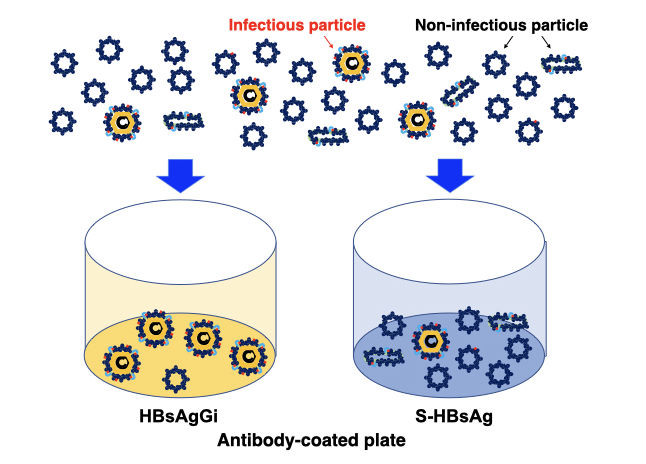
HBV virion is covered by envelope protein (HBsAg). HBsAg is modified with N- and O-glycan in subtype-specific manner (1, 2). Three types of surface antigens, S-, M-, and L-HBsAg, which are produced from an HBsAg gene containing PreS1, PreS2 and S-domains. Infection virion contain all three HBsAgs, but non-infectious particles mainly contain S-HBsAg. For the current diagnosis or analysis of HBsAg, antibodies against S-HBsAg are used to measure amount of HBsAg. RCMG’s HBsAgGi (HBsAg glycan isomer) is a monoclonal antibody specific to O-glycosylated Pre-S2 on M-HBsAg that is included in infectious HBV containing HBV DNA (3). The particles recognized by HBsAgGi differ from the particles recognized by S-HBs antibody. Patients have less infectious HBV particles than noninfectious particles and the ration of both particles differ in each patient. Thus, measuring infectious HBV particles could be useful to analyze pathological conditions of patients. For this purpose, we established HBsAgGi ELISA system.
HBsAgGi ELISA can measure HBV DNA particles
Sera of HBV patient mainly contains subviral particles over the HBV DNA particles. S-HBsAg antibody can capture all particles, while HBsAgGi capture infectious HBV particles containing O-glycosylated M-HBsAg. Measuring HBV by HBsAgGi will present distinctive result from measuring by S-HBsAg antibody.
Example
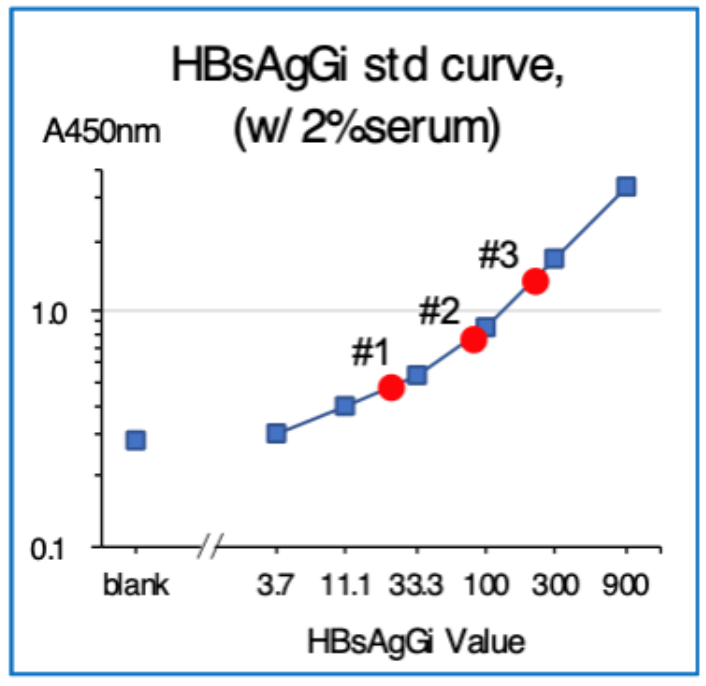
ELISA using HBsAgGi-gC coated plate and M-HBsAg as a standard sample
- Standard material: M-HBsAg (3.7 – 900 ng/mL)
- Tested samples: sera from patients with HBV (2 μL)
- Serum #1: HBsAg 6610 IU/mL, HBV DNA 3.1
- Serum #2: HBsAg 8612 IU/mL, HBV DNA 0
- Serum #3: HBsAg 36600 IU/mL, HBV DNA 8.
- Detection: Biotinylated HBsAgGi-gC antibody and HRP-labeled streptavidin
For details and ordering this service, please feel free to contact us.
References:
1. Schmitt et al. (2004) J Gen Virol 85:2045-2053.
2. Dobrica et al. (2020) Cells 9:1404.
3. Angata et al. (2021) Biochim Biophys Acta Gen Subj. 1866:130020


